Introduction
Our research utilizes human-induced pluripotent stem cells (iPSCs) to explore rare diseases, advance drug discovery, and perform toxicity assessments through iPSC-derived cellular models of rare diseases. We are also particularly interested in identifying novel biomarkers and therapeutic targets.
Keywords
Stem cells, gene editing, inherited rare diseases, disease modelling.
Research Details
Over 80% of rare diseases are genetically inherited, with most being monogenic disorders caused by defects in a single gene. Recently, induced pluripotent stem cells (iPSCs) have emerged as a powerful tool to model disease pathways and study the progression of human diseases. iPSCs are derived from somatic cells through the expression of four transcription factors, known as the "Yamanaka factors"—KLF4, MYC, OCT4, and SOX2—reprogramming them into embryonic stem cell-like pluripotent cells. This technology offers unique advantages for modeling rare diseases, as iPSCs carrying patient-specific mutations can replicate the cellular and functional phenotypes seen in affected individuals.
iPSC-based disease modeling has led to significant breakthroughs in understanding the pathogenesis of various rare disorders, contributing to clinical trials for pharmaceutical interventions and cell replacement therapies. Patient-derived iPSCs can be differentiated into specific tissues that accurately mimic patient and disease-specific phenotypes, enabling the design of personalized treatments. This approach is particularly valuable for studying the pathophysiology of neuronal and cardiac diseases and identifying novel therapeutic strategies (Figure 1).
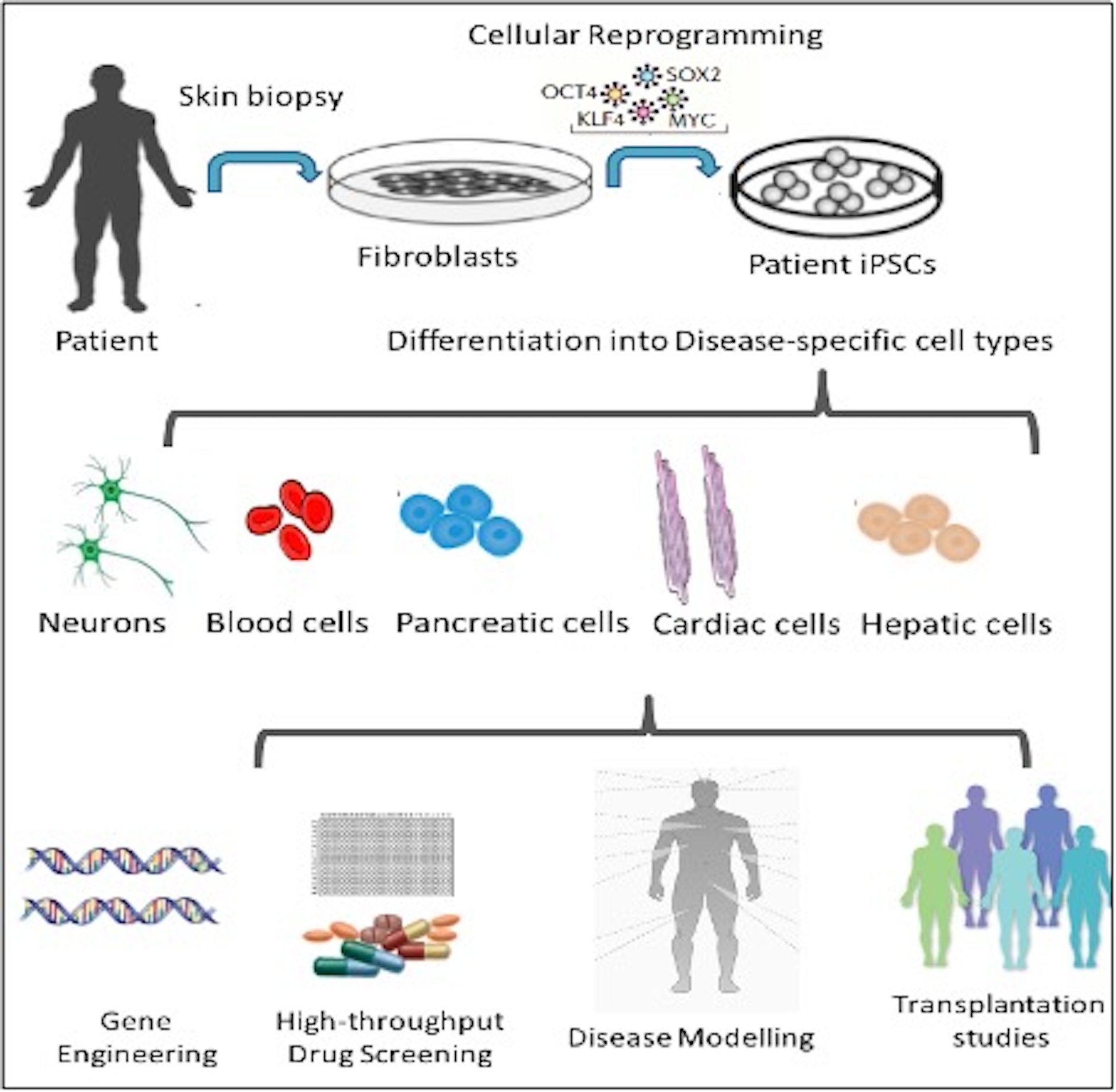
Figure 1. Potential iPSC applications in research and medicine.
Additionally, CRISPR gene editing technology has enhanced researchers' ability to model disease phenotypes by introducing or repairing mutations in differentiated cells. Our project aims to integrate advanced iPSC research with CRISPR gene editing to investigate inherited rare diseases that are prevalent in our region, largely due to the high rate of consanguinity.
In our research, we utilize the following key methodologies:
- iPSC culture and expansion: Establishing and maintaining robust iPSC lines.
- Characterization and assessment of iPSC lines: Ensuring the quality and pluripotency of the generated iPSCs.
- Developing and registering iPSC lines in the international stem cell bank "hPSCreg": Contributing to a global resource for stem cell research.
- iPSC differentiation into disease-specific cell types: Generating specialized cell types, such as neuronal, retinal, and cardiac cells, to model specific diseases.
- Genome editing using the CRISPR-Cas9 system: Introducing precise genetic modifications to study disease-causing mutations.
- Neuronal toxicity and chemotherapy testing using differentiated neurons: Assessing cellular responses to toxins and therapeutic agents in a controlled environment.
- In vitro drug screening and target validation using iPSC-differentiated cells: Identifying therapeutic targets and evaluating the efficacy of drug candidates.
Using these technologies, we have successfully developed in vitro models for inherited rare diseases and performed functional analyses to investigate disease-specific phenotypes (Figure 2).
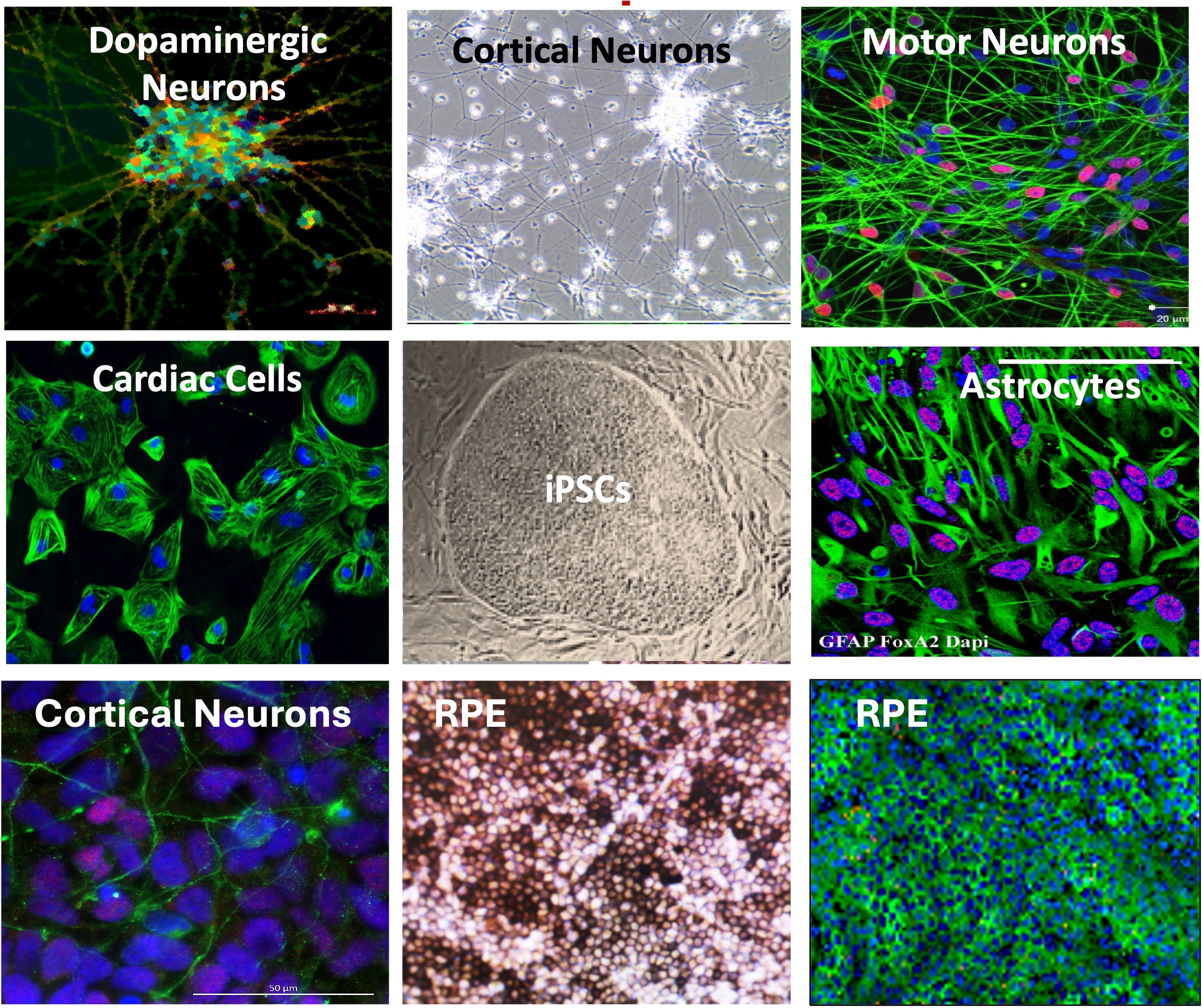

Figure 2. Differentiated cells derived from our iPSCs, including neurons and cardiac cells. These models provide significant value for studying inherited diseases that impact cells from otherwise inaccessible tissues, enabling deeper insights into disease mechanisms and potential treatments.
Impact and Significance
In our laboratory at the CTC, we generate iPSCs by delivering the four reprogramming factors to dermal fibroblasts. Additionally, we have successfully reprogrammed other cell types, such as gingival fibroblasts, into iPSCs. These iPSCs are subsequently differentiated into disease-specific cell types, allowing us to investigate the underlying mechanisms of various inherited diseases.
Key Publication
- Nidaa A Ababneh , Dema Ali, Ban Al-Kurdi, Raghda Barham, Isam K Bsisu, Deema Dababseh, Sally Arafat , Asim N Khanfar, Leen Makahleh, Abdee T Ryalat, Malik Sallam, Mohammed El-Khateeb, Basil Sharrack, Abdalla Awidi. The utility of whole-exome sequencing in accurate diagnosis of neuromuscular disorders in consanguineous families in Jordan. Clin Chim Acta. 2021 Oct 5;S0009-8981(21)00345-4.
- Nidaa A Ababneh, Raghda Barham, Ban Al-Kurdi, Dema Ali, Nour Sharar, Sabal Al Hadidi, Renata M Alatoom, Suzan Zalloum , Mohammad H Gharandouq, Leen Makahleh, Leena N Alnsour, Hebah Alshahwan, Mohammed El-Khateeb, Abdalla Awid. Generation of an induced pluripotent stem cell (iPSC) line (JUCTCi017-A) from a patient with limb-girdle muscular dystrophy (LGMD) due to a homozygous p.Lue287Ser fs14*
-Nidaa A. Ababneh, Dema Ali, Ban Al-Kurdi, Malik Sallam, Abdulla M Alzibdeh, Bareqa Salah, Abdee T Ryalat , Belal Azab, Basil Sharrack , Abdalla Awidi. Identification of APTX disease-causing mutation in two unrelated Jordanian families with cerebellar ataxia and sensitivity to DNA damaging agents. PLoS One. 2020 Aug 4;15(8):e0236808. doi: 10.1371/journal.pone.0236808. eCollection 2020.
- Nidaa A. Ababneh, Ban Al-Kurdi, Dema Ali, Duaa Abuarqoub, Raghda Barham, Abdulla M Alzibdeh, Asim N Khanfar, Ahmad M Altantawi, Abdee T Ryalat, Basil Sharrack, Abdalla Awidi. Generation and characterization of induced pluripotent stem cell (iPSC) line (JUCTCi002-A) from a patient with ataxia with oculomotor apraxia type 1 (AOA1) harboring a homozygous mutation in the APTX gene. Stem Cell Res. 2020 Jul 25;48:101925. doi: 10.1016/j.scr.2020.101925.
Collaborative Partners
Team Members Prof Abdalla Awidi Prof Amira Masri Prof Lyndon DA CRUZ Prof Basil Sharrack Dr Ahmad AL-Khleifat Dt Tareq Saleh Ban Al-Kurdi Raghda Barham Fairouz AlNairat Institutions: University College London King's College London University of Sheffield
Contact Details
Researcher Name: Nidaa Anwar Ababneh
Associate Researcher/ Cell Therapy Center
E-mail: n.ababneh@ju.edu.jo