Introduction
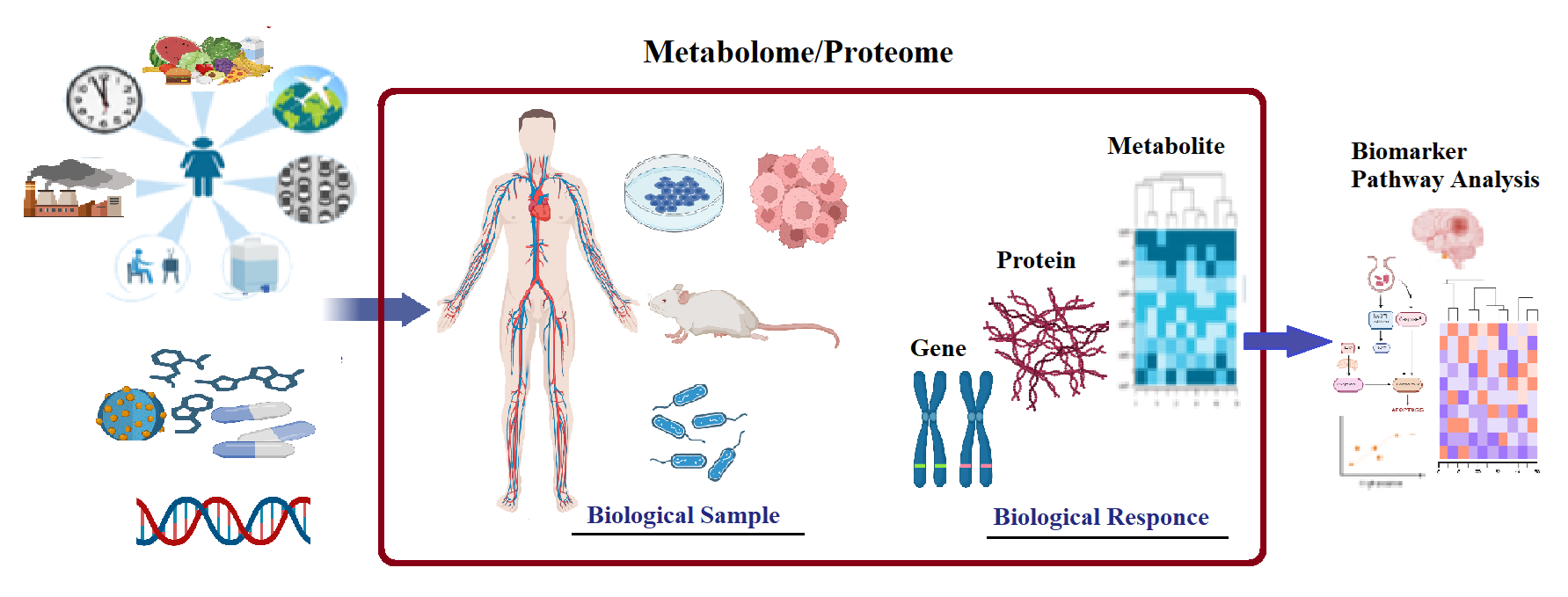
Omics primarily aims at the universal detection of genes (genomics), mRNA (transcriptomics), proteins (proteomics) and metabolites (metabolomics) in a specific biological sample. Omics studies represent advanced and promising measurement approaches for diseases and therapeutics biomarkers discovery. Currently, integrating multiple omics datasets is a promising analytical approach to provide holistic insights to disease pathophysiology, a more practical P4 medicine (predictive, preventive, personalized, and participatory) development, and tailoring more effective therapies to individual patients. Metabolomics is defined as the global identification and quantification of a set of small molecules, less than 1500 Daltons present in a specific biological system. While metabolites are the most functional biomolecules to reflect the phenotype proteomics provide a useful window into a range of biological processes and allow the identification of differentially expressed proteins between samples. Together with several international collaborators we apply state of-the-art analytical approaches including targeted and untargeted metabolomics, pharmacometabolomics, lipidomics, and proteomics to provide novel insights into pathophysiological mechanisms underlying diseases, identify unique diagnostic and prognostic biomarkers, investigate drug/treatment mechanism of action and side effects at the molecular level, and identify metabolomics surrogate pattern for drug exposome to promote health outcomes at a personalized level.
Keywords
Metabolomics, Bioanalysis, Mass Spectrometry, Biomarker, Parkinson's Disease, Cancer, Pharmacometabolomics
Research Details
In our research we focus on fundamental and applied studies in bioanalysis for biomarker discovery using various omics approaches including targeted and untargeted liquid and gas chromatography-mass spectrometry (LC/GC-MS) for metabolomics and lipidomics analyses, and LC-MS proteomics analysis. By using state of the art analytical approaches, we aim to:
- Provide novel insights into the underlying disease pathophysiological mechanisms (e.g Parkinson's Disease)
- Identify unique biomarkers for the early diagnosis and progression of disease ((e.g Parkinson's Disease)
- Investigate the pharmacological response for a treatment at the molecular level, and predict drug mechanism of action and side effects (pharmacometabolomics)
- Identify metabolomics and lipidomics surrogate pattern for drug exposome
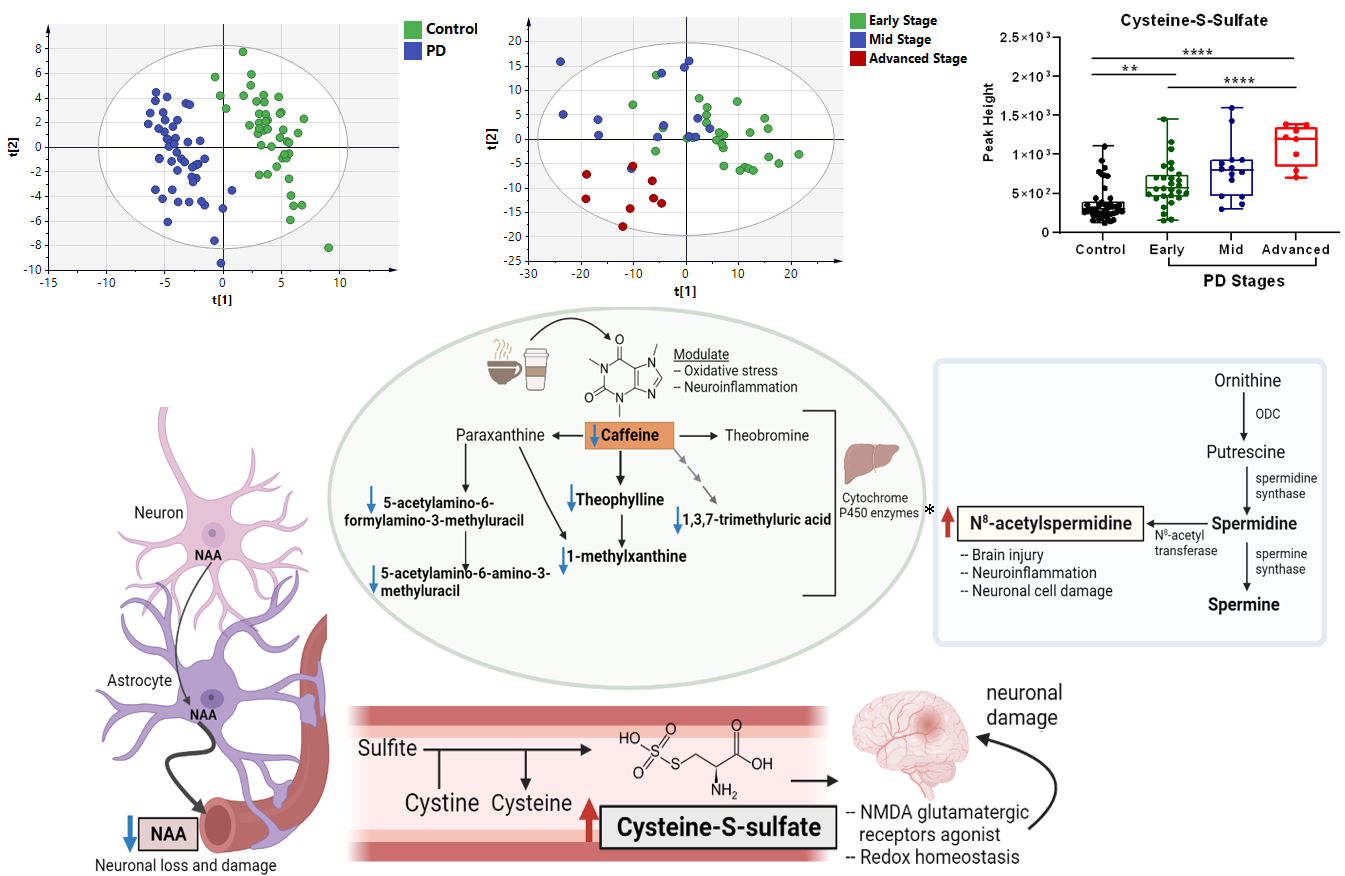
Study (1): Untargeted Serum Metabolomics and Lipidomic Revealed Potential Biomarkers for Diagnosing Parkinson's Disease and Monitoring its Progression
Parkinson's disease (PD) is a prevalent neurodegenerative disorder with a poorly understood etiology. An accurate diagnosis of idiopathic PD remains challenging as misdiagnosis is common in routine clinical practice. Therefore, identification of potential PD biomarkers and providing a better understanding of the underlying disease pathophysiology are urgent. In this study, serum samples from 50 patients with different stages of idiopathic PD (early, mid and advanced) and 45 age-matched controls were profiled using HILIC-MS/MS and GC-TOF MS based metabolomics approaches. Non-polar metabolites and lipids were profiled using reversed phase LC-MS lipidomics.
A total of 169 lipids and 212 metabolites were significantly altered in the serum of patients with PD compared to controls. Among the upregulated metabolites and lipids in PD are cysteine-S-sulfate, N-acetyl tryptophan and saturated lysophosphatidylcholines. Lower levels of N-acetylaspartic acid and ceramides (e.g. Cer 40:0, 42:0) were detected in the serum of PD patients compared to controls. Set enrichment analysis revealed a decrease in xanthines, including caffeine and its downstream metabolites, in patients with PD relative to controls and in advanced PD versus early stage PD. This suggests that caffeine and its metabolites might have a potential role in protecting against neuronal damage and decelerating PD progression among early stage PD patients. Furthermore, PD progressed from early to advanced stages with decreasing levels of LPI 20:4 and increasing levels of cysteine-S-sulfate.
The study shows an intriguing number of robust changes in specific serum lipids and metabolites that may become useful for diagnosing PD and its progression, once panels have been validated in larger clinical trials and prospective studies. Unusual metabolites like cysteine-S-sulfate and LPI 20:4 might point to therapeutic targets that could enhance the development of novel PD treatments, such as N-methyl-D-aspartate (NMDA) antagonists and GPR55 agonism, respectively.
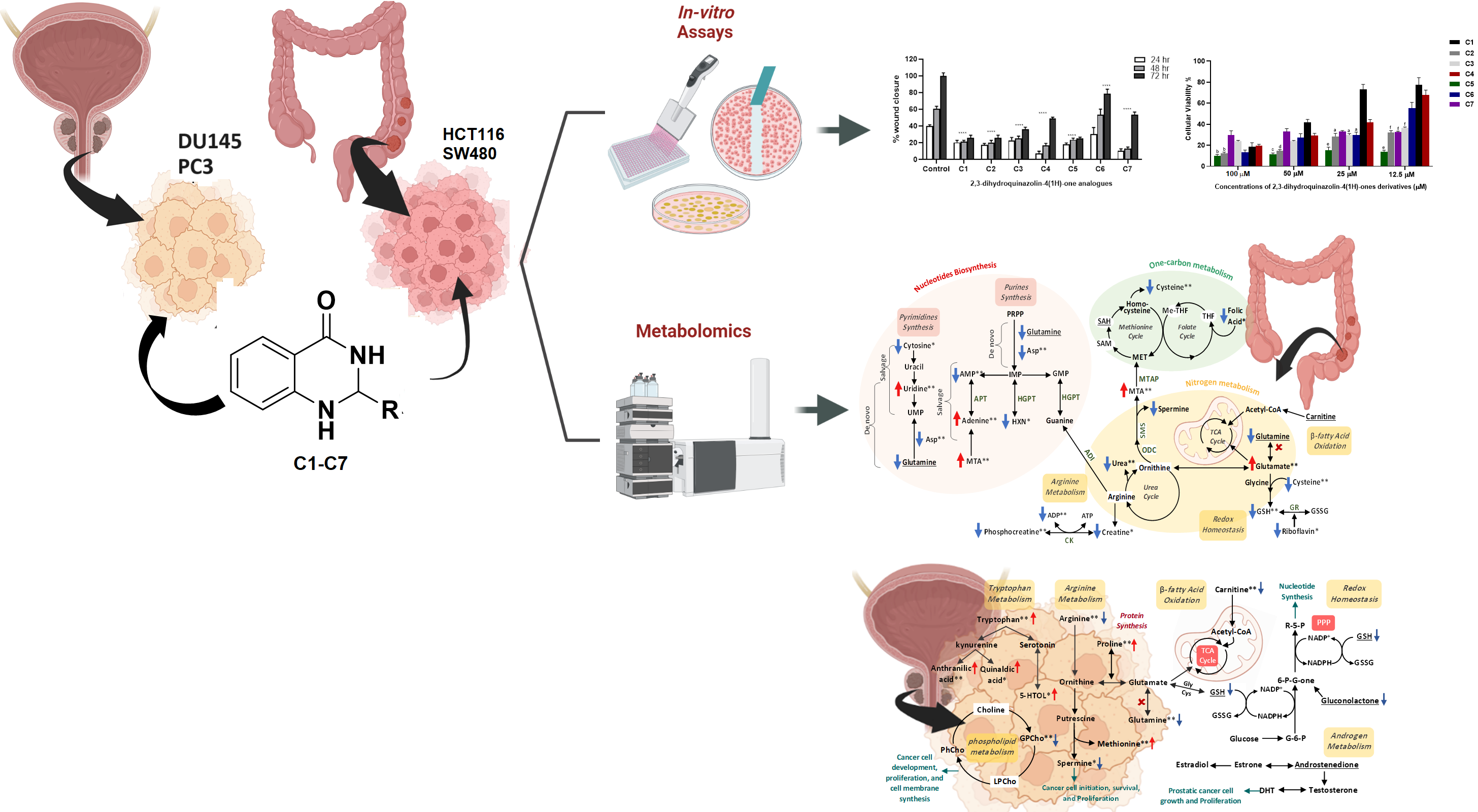
Study (2): Mass spectrometry-based metabolomics approach and in vitro assays revealed promising role of 2, 3-dihydroquinazolin-4 (1H)-one derivatives against colorectal and prostate cancer cell lines
Many of the current cancer therapies have limitations due to multidrug resistance and/or severe side effects. Quinazoline derivatives are promising lead compounds with a wide range of pharmacological actions. In this study, the effect of seven synthesized 2,3-dihydroquinazolin-4(1H)-one analogues as potential anticancer agents against two colorectal and prostate cancer cell lines was investigated using cell viability proliferation assay followed by migration, adhesion and invasion assays. A liquid chromatography-mass spectrometry (LC-MS/MS) metabolomics approach was used to identify the underlying biochemical pathways disturbed in dihydroquinazoline treated-cells.
Four of the seven compounds showed promising antiproliferative effects. Metabolite profiling of cancer cells treated with the most potent compound (IC50 < 5 µM) pointed to a significant effect of the compound on biochemical processes essential for cell proliferation and progression including amino acids biosynthesis and metabolism, redox homeostasis, purine and pyrimidine metabolism, energy generation and fatty acid oxidation. Our findings indicate that 2,3-dihydroquinazolin-4(1H)-one analogues, particularly C5, could act as promising anticancer agents, and shed a light on the fundamental role of LC-MS metabolomics in identifying new therapeutic targets and providing a better understanding of the underlying mechanisms altered in treated cancer cells.
 Publications and Links
Publications and Links
- Diagnosing Parkinson's Disease and Monitoring its Progression: Biomarkers from Combined GC-TOF MS and LC-MS/MS Untargeted Metabolomics. 2024 Doi: https://doi.org/10.1016/j.heliyon.2024.e30452
- Serum-Based Lipid Panels for Diagnosis of Idiopathic Parkinson's Disease. 2023 Doi: https://doi.org/10.3390/metabo13090990
- Mass spectrometry-based metabolomics approach and in vitro assays revealed promising role of 2, 3-dihydroquinazolin-4 (1H)-one derivatives against colorectal cancer cell lines, 2023 Doi: https://doi.org/10.1016/j.ejps.2023.106378
- Molecular and metabolic alterations of 2,3-dihydroquinazolin-4(1H)-one derivatives in prostate cancer cell lines, 2022 Doi: https://doi.org/10.1038/s41598-022-26148-4
Research Group Website: https://research.ju.edu.jo/research/groups/BBD/home.aspx
Researcher Names
- Lina A. Dahabiyeh; Associate Professor of Bioanalysis, School of Pharmacy, The University of Jordan, Jordan
- Refat Nimer; Assistant Professor, Department of Medical Laboratory Sciences, JUST, Jordan
- Yasser Al-Bustanji; Professor, Collage of Medicine, University of Sharjah, UAE
- Mohammad H. Semreen; Professor, Collage of Pharmacy, University of Sharjah, UAE
- Oliver Fiehn, Professor, West Coast Metabolomics Center, University of California, Davis, USA
Contact Information
Dr. Lina A. Dahabiyeh, PhD, FWIS IRS, Fulbrighter
Associate Professor of Bioanalysis and Pharmaceutical Analysis
Department of Pharmaceutical Sciences
School of Pharmacy
The University of Jordan
Amman 11942
Tel: 00962 5355000 Ext.23306
Email: L.dahabiyeh@ju.edu.jo